10.31047/1668.298x.v41.n1.41895
Artículos
Selection of bacterial isolates with fungal inhibition against alfalfa phytopathogens to constitute a bacterial consortium
Selección de cepas bacterianas con capacidad antifúngica contra fitopatógenos de alfalfa para construir un consorcio bacteriano
M V Moreno 1
V Arolfo 1
J López 1
S Erdozain 1
E D Bigatton 1
I Ayoub 1
E Lucini 1
A Lagares 1
A Odorizzi 1
1 Moreno, M. V/. (ORCID: 0000-0002-9699-0349), Arolfo, V/. (ORCID: 0000-0001-9514-7977), Odorizzi, A. (ORCID: 0009-0001-2375-9461), Instituto Nacional de Tecnología Agropecuaria, Estación Experimental Agropecuaria Manfredi. Manfredi, Córdoba, Argentina. Erdozain, S. (ORCID: 0009-0001-8058-6691), Lagares, A. (ORCID: 0000-00032644-8324), Universidad Nacional de La Plata, Facultad de Ciencias Exactas. La Plata, Buenos Aires, Argentina. López, J. (ORCID: 00000002-6942-7032), Universidad Nacional de La Platel, Facultad de Ciencias Exactas. La Plata, Buenos Aires, Argentina. Consejo Nacional de Investigaciones Científicas (CONICET), Argentina. Bigatton, E. D. (ORCID: 0000-0002-7896-5369), Ayoub, I. (ORCID: 0000-0001-8286-108X), Consejo Nacional de Investigaciones Científicas (CONICET), Argentina. Universidad Nacional de Córdoba, Facultad de Ciencias Agropecuarias. Córdoba, Argentina. Lucini, E. I. (ORCID: 0000-00030385-3271), Universidad Nacional de Córdoba, Facultad de Ciencias Agropecuarias. Córdoba, Argentina. Correspondence to: moreno.maria@inta.gob.ar
ABSTRACT
Alfalfa breeding programs require environmentally friendly alternatives to improve plant growth and health. Plant Growth Promoting Rhizobacteria (PGPR) and endophytic bacteria offer a biological solution as they can inhibit phytopathogenic fungi by producing antifungal compounds or enzymes. In Argentina, alfalfa is primarily affected by Colletotrchum trifolii (Ct) and Phytophthora megasperma (Pm). The objectives of this study were to select bacterial isolates with antifungal inhibition against Ct and Pm and identify a biocontroller isolate compatible with the rhizobia INTA5 has and with low-N2O emissions, to constitute an eco-friendly bacterial consortium. Ten isolates demonstrated antifungal inhibition, with mean Inhibition Percentage (IP) values between 41.74-91.28 for Ct and 56.23-77.59 for Pm compared to the control. Among these isolates, B2, B4, SC6, and MN1 showed the highest inhibition performance and were selected to evaluate their compatibility with INTA5. Only B4 exhibited compatibility, which was further evaluated against Ct in an experiment with alfalfa seedlings under controlled conditions. The alfalfa seedlings treated with B4 increased the plant survival number against Ct. The availability of a bacterial consortium that promotes biocontrol and biological nitrogen fixation represents the preferred strategy for alfalfa breeding programs.
Keywords: Medicago sativa L., biological control, breeding strategies, endophytes, plant growth promoting rhizobacteria (PGPR).
RESUMEN
El programa de mejoramiento de alfalfa requiere alternativas más amigables con el ambiente para incrementar la sanidad del cultivo. Las rlzobacterlas promotoras del crecimiento vegetal (RPCV) y endófltos de semillas constituyen una opción para inhibir hongos patógenos por la producción de compuestos antifúngicos o enzimas hidrolíticas. En Argentina, la alfalfa es afectada principalmente por Colletotr¡chum trifolii (Ct) y Phytophthora megasperma (Pm). Los objetivos fueron seleccionar bacterias con capacidad antifúngica contra Ct y Pm, e identificar una compatible con el rizobio INTA5 con baja emisión de N2O para constituir un consorcio bacteriano sustentable. Diez cepas mostraron capacidad antifúngica con medias de porcentaje de inhibición (PI) entre 41,74-91,28 para Ct y 56,23-77,59 relativos al control para Pm. B2, B4, SC6 y MN1 fueron seleccionadas por su mayor capacidad de biocontrol y se estudió la compatibilidad con INTA5. Sólo B4 resultó compatible, por ello fue evaluada contra Ct sobre plántulas de alfalfa en experimentos bajo condiciones controladas. Esta cepa incrementó el número de plántulas sobrevivientes cuando las mismas fueron inoculadas con Ct. La disponibilidad de un consorcio bacteriano que estimule tanto el biocontrol como la fijación biológica de nitrógeno representa la estrategia preferida por el programa de mejoramiento de alfalfa.
Palabras clave: Medicago sativa L., control biológico, mejoramiento, endófitos, rizobacterias promotoras del crecimiento vegetal (RPCV).
INTRODUCTION
Alfalfa (Medicago sativa L.) is a perennial forage legume of worldwide importance. It is a high-yielding and easily digested animal fodder with high nutritional value (Noori et al., 2018). In Argentina, alfalfa cultivation represents annually 3.2 million ha and is mainly used as animal forage (Basigalup et al., 2020). It is widely spread across the production areas and could be affected by almost twenty diseases with different degrees of importance (Gieco et al., 2007). Two of the most critical fungal diseases, due to their severity and frequency of appearance in the Pampas Region, are anthracnose caused by Colletotrichum trifolii (Ct) and root rot caused by Phytophthora megasperma (Pm). Biocontrollers obtained from microorganisms are a sustainable alternative for the inhibition of phytopathogens to reduce the use of agrochemicals and their negative impact on the environment (Qiao et al., 2014).
During the last three decades, many researchers reported the presence of seed endophytes in several plant species (Díaz Herrera et al., 2016). Seed endophytes may come from different plant organs and are transferred to seeds via vascular connections or through gametes, resulting in colonization of the embryo and endosperm (Malfanova et al., 2013). After seed germination, these populations increase and colonize different tissues, including roots, reaching the endorhizosphere and probably also the exorhizosphere (Hardoim et al., 2012; López-López et al., 2010). The role of seed endophytes has not been unraveled yet. It has been demonstrated that they can promote plant growth by encouraging hormone production or by enhancing nutrient acquisition, especially nitrogen and phosphorus (Gagne-Bourgue et al., 2013; Xu et al., 2014). On the other hand, the antifungal activity of several bacterial seed endophytes (such as Baclllus and Pseudomonas) has also been recognized. These microorganisms could produce lipopeptides such as surfactin, iturin, and mycobacillin with antifungal activity (Gagne-Bourgue et al., 2013). Antifungal activity by seed endophytes was recorded against different Fusarlum species like F. oxysporum which causes wilt in tomatos (Sundaramoorthy and Balabaskar, 2013) and rice (Mukhopadhyay et al., 1996) and F. gramlnearum that is the causative agent of wheat head blight (Díaz Herrera et al., 2016) . The microbiome in alfalfa plants has been recently explored in leaves, stems, and root nodules to investigate correlations between the soil bacterial community and the role of endophytes (Pini et al., 2012). Recent reports characterize the diversity and main phenotypic traits of the seed microbiome in alfalfa. López et al. (2018) revealed that Baclllus genera (approximately 13 different Baclllus spp. strains), among other 40 genera, presented fungal inhibition mechanisms against Sclerotlnla sclerotlorum.
Likewise, there are plant growth-promoting rhizobacteria (PGPR) exerting their ¡nfluence on crop growth and health through diverse mechanisms, such as phytohormone production, phosphate solubilization, nitrogen fixation, and biological control of pathogens. These microorganisms are also classified according to their functional activity: I) biofertilizers (increase the availability of nutrients to the plant), II) phytostimulators (promote growth through the production of phytohormones), III) rhizo-remediators (contribute to the degradation of polluting compounds), IV) biopesticides (control diseases) (Ahemad and Kibret, 2014). In order to design desirable bioproducts it is recommended to combine two or more of these mechanisms (Rojas Badía et al., 2017).
Alfalfa is the main source of vegetable protein in beef cattle and dairy production due to its ability to acquire large amounts of nitrogen by symbiosis (biological nitrogen fixation: BNF) (Jozefkowicz et al., 2017) . Alfalfa cultivation, also incorporates nitrogen fixed to the soil, thus improving the production of non-legumes during crop rotation (Jozefkowicz et al., 2017). Different nitrogen-fixing alfalfa symbionts originating from different ecoregions were screened for low N2O emissions, and novel rhizobia was reported (Slnorhlzoblum melllotl INTA1-6). The commercial strain Slnorhlzoblum melllotl B399 has also conserved nitrate (NAP+), nitrite (NIR+), and nitric oxide (NOR+) gene clusters related to nitrous oxide (N2O) production from nitrate. Brambilla et al. (2019) reported these nitrogen-fixing alfalfa symbionts which exhibit the nitrous oxide reductase gene (NOS+) and demonstrate exceptionally low N2O emissions. Mutations were only detected in the structural gene for nitrate reductase (napC) in the denitrification cluster. No significant differences in plant productivity or nodule number were observed when plants were inoculated with the B399 and the INTA4-6 strains, suggesting that low-N2O-emitting rhizobia can be an ecological alternative to the current inoculants without refusing the economic profitability (Brambilla et al., 2019).
A bacterial consortium that stimulates biocontrol and BNF represents the preferred strategy to improve alfalfa production. This study aims to select bacterial isolates with antifungal inhibition against Ct and Pm alfalfa phytopathogens and select a biocontroller strain compatible with the native rhizobia INTA5 with low-N2O emissions to constitute an eco-friendly bacterial consortium for alfalfa crop.
MATERIALS AND METHODS
In-vitro antagonistic experiments
Ten PGPR rhizosphere isolates (PSE7, PSE, RI1, RI9, SC1, SC2, SC6, MN1, SI2, EZE) provided by the Faculty of Agricultural Sciences, National University of Córdoba (Bigatton et al., 2021) and four endophytic alfalfa isolates (B2, B4, B12, B22) isolated by López et al. (2018) and provided by the Faculty of Exact Sciences, National University of La Plata were evaluated. Most of the isolates evaluated in this work (85.7 %) belong to the Baclllus genus; only PSE7 and PSE are Pseudomonas genus. Bacterial isolates were grown in tryptic soy broth (TSB) for 48 h at 28 °C. After growth, each isolate was serially diluted in saline solution (SS) to quantify their population density by the spread plate method. The bacterial number was adjusted at 108 colony-forming units (CFU).mL-1. Phytopathogenic fungus Colletotrlchum trlfolll (Ct) was provided by Dr. N. Bernardi (IPAVE-INTA), and Phytophthora megasperma (Pm) was isolated from an infected plot at INTA-EEA Manfredi. A plug of each fungus was placed at the center of a petri dish with potato-dextrose-agar (PDA). A bacterial stretch was placed on each side of the plug (Astorga-Quirós et al., 2014), and plates were incubated for 7-15 days at 28 °C. Negative control plates with fungal plugs without bacterial stretch were used. Each experiment had three repetitions. Fungal growth inhibition was considered positive when mycelial growth was inhibited around a bacterial stretch on antagonistic plates. For evaluation, fungal colony diameter was measured in the presence of bacterial stretches or antagonistic plates and control plates. Inhibition percentage (IP) was calculated according to Rojas-Badía et al. (2017). Statistical analysis was made using the ANOVA model and Tukey test for the mean comparisons. This analysis was carried out using the InfoStat software (Di Rienzo et al., 2018).
Bacterial compatibility experimentsTo perform compatibility experiments, the rhizobial isolate INTA5 was grown in trypticase soy broth (TSB) for 24 h at 28-30 °C (reached 108 CFU. mL-1). A 100 pL aliquot of this culture was seeded on a Petri dish containing nutritive agar (NA) (adapted from Burgos-Toro, 2019). Petri dishes were then dried for 40 min before 1 cm sterile Whatman filter paper discs were placed on them. 5 pL of each biocontrol isolate was inoculated on every paper and were incubated for 48 h at 28-30 °C. Each experiment had three replicates. The absence of an inhibition halo around the biocontrol colony was evidence of strain compatibility (Díaz Herrera et al., 2016).
Antagonistic experiments in seedlings under controlled conditionsThe most effective biocontroller B4 was grown in TSB and incubated at 28 °C for 48 h. After incubation it was diluted in half with 5 % sterile saccharose solution to improve the adhesion of the seed surface. Alfalfa seeds were sterilized with 2 % NaClO for 3 min, 75 % ethyl alcohol for 4 min, and rinsed three times with sterilized water. In the last wash, disinfection control was carried out. The cultivars Amaya PV INTA (A), and Costera INTA (C), which are moderately resistant and susceptible to Ct, were used (Basigalup et al., 2020; Odorizzi, 2015). The treatments were CE (only with B4), T (with B4 and Ct), and CP (only with Ct). One-third of the seeds of each variety (A and C) were not inoculated with the endophyte (CP treatment). The remaining seeds were inoculated with B4 (CE and
T treatments) and were dried for 1 h. The seeds were sown in nine plástic trays filled with moist sterilized soil, in two rows per variety for each tray with three replicates per treatment. Experiments were performed with three replicates. Trays were placed randomly in an artificial climate chamber at 25 °C with light (16 h)/darkness (8 h). After ten days, the seedlings of CP and T trays were uniformly sprayed with Ct spore suspension with a concentration of 2.106 spores.mL-1. Spore count was performed in a New Bauer chamber. Before inoculation, the number of emerging seedlings was recorded. Then, the trays were placed in a humidity chamber at 23 °C for two days to favor the stomata opening and the phytopathogen infection. The count of surviving seedlings was evaluated fifteen days after inoculation. The adjusted model corresponded to a binomial distribution since the response variable is a proportion, using the AIC criterion. This analysis was conducted using the Generalized Linear Mixed Model (GLMM) module with the InfoStat software (Di Rienzo et al., 2018). The fixed variables were treatments (CE, T, CP) and genotypes (A, C), while blocks and trays were considered random variables. To achieve independence and robustness in the analysis, trays were nested in blocks and treatments (Figure 1).
RESULTS
In-vitro antagonistic experiments
Out of all the strains, nearly ten, including both PGPR and endophytes, demonstrated inhibition of pathogens. They showed different levels of IP against Ct at fifteen days between 41.74 and 91.28 (Figure 2 A and 2 B, Table 1). Only four isolates did not show inhibition for one or both phytopathogens (B12, PSE7, PSE, and SI2). Significant differences against Ct were observed between B2, B4, and B22 (p<0.0001). IP values for Pm showed significant differences (p<0.0001) and less variation than Ct, between 56.23 and 77.59 (Figure 2 C and 2 D, Table 1).
Considering that the IP values in Ct showed high variation, we selected the isolates based principally on this pattern because with Pm all isolates, with the only exception of PSE, had an acceptable antifungal effect (IP>50). The IP threshold for Ct was established at 54 according to references (Rojas-Badía et al., 2017; Guo et al., 2020). Based on the results, two endophytic (B2, B4) and two PGPR isolates (SC6, MN1) were selected to evaluate in bacterial compatibility experiments.
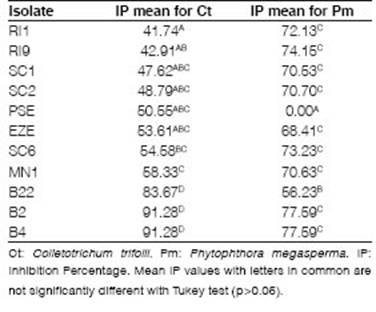
Table 1: IP mean valúes of bacterial isolates evaluated for both phytopathogens (Ct and Pm).
Bacterial compatibility and antagonistic experi-ments in seedlings under controlled conditionsB4 strain was the only isolate compatible with natural alfalfa rhizobia INTA5 (Figure 3). B4 was used for evaluation in controlled conditions for antagonistic experiments.
Seeds treated with B4 (T) had a higher number of surviving seedlings than those treated with phytopathogen control (CP) (Table 2). The symptoms observed were yellowing of leaves and Progressive senescence. There was no significant reduction in the proportions of surviving plants for endophytic control (CE) with Amaya PV INTA (A) (0.89) and Costera INTA (C) (0.86) alfalfa varieties. These results could indicate that the endophytic isolate does not affect the seed germination or the normal development of the seedlings during the test. When the interaction term is significant, the significances of the factors alone are not relevant. The treatment x genotype interaction (p=0.0026) showed significant differences, as well as between treatments and genotypes (p<0.0001). In both genotypes, B4 improves the behavior of the seedlings in the presence of Ct (trays T), showing a protective effect. This finding confirms the fungal inhibition of B4 on Ct in the host-phytopathogen-biocontroller biological system. Inoculation with the endophyte had a greater improvement in genotype C (0.12) than in genotype A (0.09) when they were confronted with Ct.
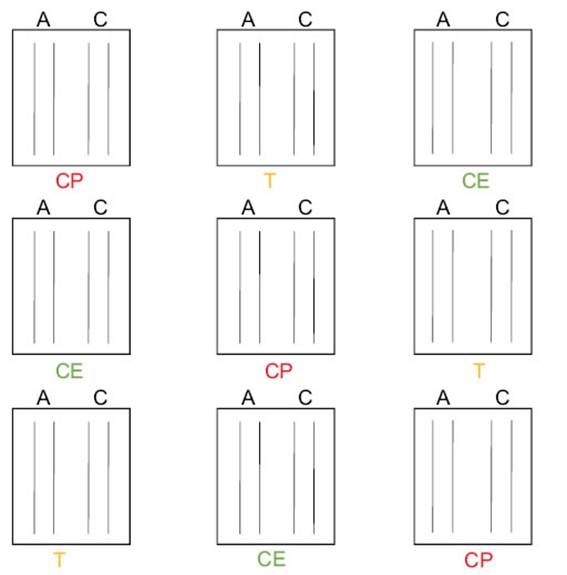
Figure 1: Experimental design of antagonistic experiments under controlled conditions.
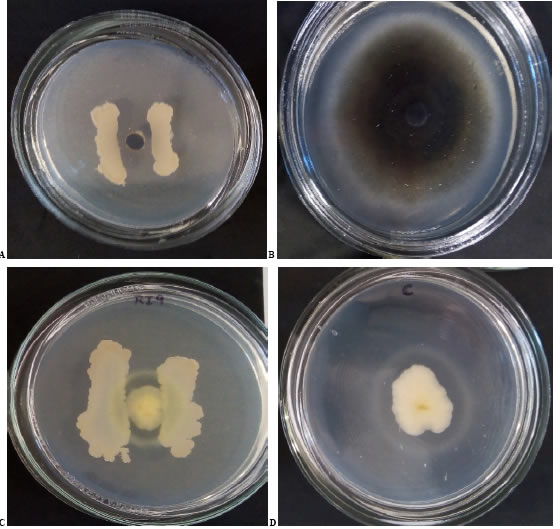
Figure 2: Photographs of isolates against Ct and Pm at 15 days of incubation.
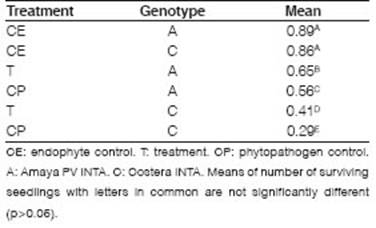
Table 2: Adjusted mean of proportions of surviving plants for treatment x genotype interaction evaluated in controlled conditions experiments.
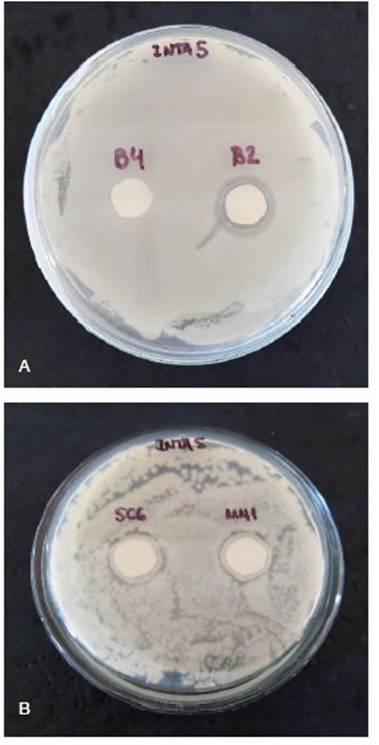
Figure 3: Photographs of biocontrol isolates over INTA5 at two days of incubation. A: B2 and B4 (endophytic). B: SC6 and MN1 (rhizospheric). No halo was present in the B4 isolate.
DISCUSSION
PGPR and endophyte isolates had shown a great result as biocontrollers of alfalfa phytopathogens. Significant differences between B2, B4, and B22 with remaining isolates against Ct could be associated with the fact that they are endophytic strains isolated from alfalfa seeds (López et al., 2018) and may be evidence of phytopathogen speciflcity. The mean IP valúes obtained were higher than 41.74 for Ct and 56.23 for Pm (Table 1) according to the lower IP limit reported (50) for good biocontrol candidates (Astorga-Quirós et al., 2014). The highest IP values obtained agreed with Rojas-Badía et al. (2017) who reported eighty for Baclllus strains against Fusarlum oxysporum and values of fifty when the phytopathogen was Fusarlum monlllforme. The different IP ranges recorded for Ct and Pm indicated distinct biocontrol capabilities of each microorganism. These results suggest that each isolate could produce metabolites with diverse specificity. This situation was reported for Baclllus strains (Rodríguez et al., 2016; Rojas-Badía et al., 2017; Fathi et al., 2018). Variations in the biocontrol potential of an isolate depend on genetic characteristics, environmental factors, and interactions with other microorganisms (Astorga-Quirós et al., 2014).
Most of the isolates evaluated in this work (85.7 % of the total isolates) belong to the Baclllus genus which is characterized by the production of diverse metabolites like antibiotics, lipopeptides, or lytic enzymes (Rojas-Badía et al., 2017, López et al., 2018). Our results coincide with previous reports using Baclllus subtllls strains on alfalfa seedlings against Colletotrlchum trlfolll under controlled conditions. A reduction in the incidence and severity of illness was observed due to the decrease in the conidia germination and the lysis of some of them (Douville and Boland, 1992). Likewise, it was recently discovered that the growth of Phytophthora nlcotlanae (tobacco pathogen) was suppressed by peptides and proteins produced by Baclllus velezensls, with irreversible damage to the cell wall and membrane (Guo et al., 2020). Likewise, Baclllus amylollquefaclens showed biocontrol capacity against this patogen (Guo et al., 2019).
Pseudomonas genus had biocontrol activity against Ct (Yu et al., 2022). PSE strain demonstrated biocontrol efficacy against Ct, consistent with characteristics of the genus. Accordingly, Pseudomonas aureofaclens reduces the infection index and severity of the Phytophthora megasperma fungus in Asparagus officinalis seedlings. Plant performance is improved through growth stimulation and the antibiotic capacity of the biocontrol strain (Carruthers et al., 1995). Similarly, Pseudomonas fluorescens showed high levels of enzymatic activity and phenol production against Phytophthora drechslerl in Pistacia vera (Fathi et al., 2018).
Combining multiple PGPR strains is crucial for meeting the diverse nutrient requirements of crops in modern agriculture, instead of relying on a single strain and function fertilizer (Li et al., 2020). In this sense, a bacterial consortium is a technological tool for crop sustainability. In our study, only the B4 isolate was compatible with the native alfalfa rhizobia INTA5 (Figure 3). This finding is significant for environmental sustainability because INTA5 is a nitrogen-fixing alfalfa symbiont native from Córdoba and exhibits exceptionally low N2O emissions (Brambilla et al., 2019). These authors have reported the presence of N2O reductase genes (NOS+) that significantly reduce these emissions at < 0.1 mg N2O.kg soil-1. No significant differences in plant productivity or nodule number were observed between the commercial strain B399 and strains INTA4-6, suggesting that at least a subgroup of the eco-friendly rhizobia can replace current high N2O-emitting alfalfa inoculants without economic penalties (Brambilla et al., 2019).
Since the number of surviving seedlings in the T trays was significantly higher than in the CP trays (Table 2), it can be concluded that seed inoculation with B4 before sowing had a protective effect on seedlings against Ct. It is inferred that PGPR in the plant rhizosphere and seed endophytes could induce systemic resistance in stems and leaves on crops, by producing volatile organic compounds, lipopolysaccharides, siderophores, and stress-related plant hormones (e.g., abscisic acid, jasmonic acid, and ethylene) (Ruzzi and Aroca, 2015; Gouda et al., 2018; Yu et al., 2022). Considering these, we could induce that the B4 strain improves seedling behavior in the presence of this phytopathogen through induced systemic resistance (ISR). The behavior of A and C was as expected according to previous results of the working group (Basigalup et al., 2020; Odorizzi, 2015). Seed treatment with endophytes could be used as a technological tool for alfalfa because seed germination and seedling growth in CE trays were not affected. There was no significant reduction in the number of surviving seedlings for CE trays (0.89 for A and 0.86 for C, Table 2, Figure 4). The B4 isolate showed a high antifungal capacity in-vitro against Sclerotinia sclerotiorum, nonetheless, B2, B12, and B22 demonstrate high performance. These strains combined several hydrolytic activities (proteases, amylases, pectinases, and cellulases) with fungal inhibitory capacity. In our work, we selected the B4 isolate as the most efficient biocontroller for both pathogens (Ct and Pm) and compatible with the native alfalfa rhizobia INTA5. B4 strain may be used in a bacterial consortium formulation in the future to improve the crop performance in the fields.
The advantage of a bacterial consortium including seed endophytic strains in seed inoculation relies on the assumption that they may establish a closer association with the host plant compared to isolates from other sources, constituting the endogenous alfalfa microbiome along with other rhizosphere microorganisms. In addition, seed-associated endophytes could constitute a conserved part of a long-term hereditary consortium, facilitating the incorporation of new microbial species into the consortium and its consequent vertical inheritance (López et al., 2018). Field tests will be carried out to evaluate bacterial consortium B4-INTA5 in terms of its biocontrol capacity and the efficiency in the biological fixation of nitrogen in alfalfa varieties from the INTA EEA-Manfredi breeding program. These results will allow us to evaluate the relative plant-colonization efficiency between the bacterial consortium and the microbial rhizospheric community, as well as the productivity and performance of alfalfa varieties.
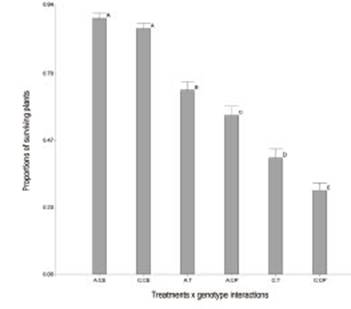
Figure 4: Adjusted mean of treatments x genotype interactions of proportions of surviving plants evaluated. Amaya PV INTA x control endophyte (A:CE), Costera INTA x control endophyte (C:CE), Amaya PV INTA x treatment (A:T), Amaya PV INTA x control phytopathogen (A:CP), Costera INTA x treatments (C:T), Costera INTA x control phytopathogen (C:CP). Means of number of surviving seedlings with letters in common are not significantly different (p>0.05).
CONCLUSIONS
B4 endophyte isolate showed the highest inhibi-tion performance against alfalfa phytopathogens and was compatible with eco-friendly native rhizobia INTA5 to constitute a consortium. The availability of a bacterial consortium that stimulates biocontrol and biological nitrogen fixation represents the preferred strategy for the alfalfa breeding program.
ACKNOWLEDGMENTS
We are grateful to Dr. Daniel H. Basigalup (INTA-EEA Manfredi: Estación Experimental Agropecuaria Manfredi) for his valuable suggestions and guidance. We also thank Dr. Nelson Bernardi (INTA-CIAP: Centro de Investigaciones Agropecuarias) for providing the Colletotrlchum trifolii isolates. This work was supported by INTA (Instituto Nacional de Tecnología Agropecuaria) with Structural Project PE 142 INTA (2019-2022).
Data availability statementThe data that support the findings of this study are available from the corresponding author upon reasonable request.
Fecha de recepción: 23/07/2023
fecha de aceptación: 12/04/2024
REFERENCES
Ahemad, M. and Kibret, M. (2014). Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. Journal of King Saud University-Science, 26, 1-20. https://doi.ora/10.1016/¡.jksus.2013.05.001
Astorga-Quirós, K., Meneses-Montero, K., Zúñiga-Vega, C., Brenes-Madriz, J. and Rivera-Méndez, W. (2014). Evaluación del antagonismo de Trichoderma sp. y Bacillus subtilis contra tres patógenos del a¡o. Tecnología en Marcha, 27(2), 82-91. https://www. scielo.sa.cr/pdf/tem/v27n2/a08v27n2.pdf Basigalup, D., Odorizzi, A. and Arolfo, V. (2020). Alfalfa (Medicago sativa L.) en Argentina. Editorial Académica Española.
Bigatton, E. D., Ayoub, I., Palmero, F., Berdini, A., Baldessari, J. J., Castillejo Sánchez, M. A., Lucini, E. I. and Haro, R. J. (2021). Plant growth promoting rhizobacteria: effects on root growth and yield of peanut (Arachis hypogaea L.) crop. South American Science, 2, e21126. https://doi.org/10.52755/sas.v2iedesp1.126
Brambilla, S., Soto, G., Odorizzi, A., Arolfo, V., McCormick, W., Primo, E., Giordano, W., Jozefkowicz, C. and Ayub, N. (2019). Spontaneous mutations in the nitrate reductase gene napC drive the emergence of eco-friendly low-N2O-emitting alfalfa rhizobia in regions with different climates. Microbial Ecology, 79, 10441053. https://doi.org/10.1007/s00248-019-01473-w
Burgos-Toro, A. D. (2019). Antagonismo e inhibición de la comunicación bacteriana en bacterias cultivables aisladas de esponjas del caribe colombiano con biofouling y sin biofouling. Unpublished master thesis, Universidad Nacional de Colombia, Colombia. http://repositorio.unal.edu. co/bitstream/handle/unal/78125/1147687914.2020.pdf?sequence=1&isAllowed=y
Carruthers, F. L., Shum-Thomas, T., Conner, A. J. and Mahanty, H. K. (1995). The significance of antibiotic production by Pseudomonas aureofaciens PA 147-2 for biological control of Phytophthora megasperma root rot of asparagus. Plant and Soil, 170, 339-344. https://doi.org/10.1007/BF00010487
Díaz Herrera, S., Grossi, C., Zawoznik, M. and Groppa, M. D. (2016). Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiological Research, 186-187, 37-43. http://dx.doi.orq/10.1016/¡.micres.2016.03.002
Di Rienzo, J. A., Casanoves, F, Balzarini, M. G., González, L., Tablada, M. and Robledo, C. W. (2018). InfoStat versión 2018. Facultad de Ciencias Agropecuarias, Universidad Nacional de Córdoba, Argentina. http:// www.infostat.com.ar
Douville, Y. and Boland, J. G. (1992). A note on the antibiotic properties of Bacillus subtilis against Colletotrichum trifolii. Phytoprotection, 73(1), 1-36. https://doi.org/10.7202/706018ar
Fathi, F., Saberi-Riseh, R. and Moradi, M. (2018). The effects of biocontrol Bacillus and Pseudomonas strains on plant growth and biochemical defense mechanisms in pistachio seedlings inoculated with Phytophthora drechsleri. Pistachio and Health Journal, 1(3), 15-26. https://doi.org/10.22123/phj.2018.144287.1010
Gagne-Bourgue, F., Aliferis, K. A., Seguin, P., Rani, M., Samson, R. and Jabaji, S. (2013). Isolation and characterization of indigenous endophytic bacteria associated with leaves of switchgrass (Panicum virgatum L.) cultivars. Journal of Applied Microbiology 114(3), 836-853. https://doi.org/10.1111/jam.12088
Gieco, J., Moreno, M. V. and Basigalup, D. (2007). Enfermedades de la alfalfa y abordaje molecular de la selección por resistencia. In: D. Basigalup (Ed.), El cultivo de la alfalfa en la Argentina (pp. 451-475). Ediciones INTA.
Gouda, S., Kerry, R. G., Das, G., Paramithiotis, S., Shin, H. S. and Patra, J. K. (2018). Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiological Research, 206, 131-140. http://doi.ora/10.1016/¡.micres.2017.08.016
Guo, D., Yuan, C., Luo, Y., Chen, Y., Lu, M., Chen, G., Ren, G., Cui, C., Zhang, J. and An, D. (2019). Biocontrol of PGPR strain Bacillus amyloliquefaciens Ba168 against Phytophthora nicotianae on tobacco. BioRxiv, 1-21. https-//do¡ org/10 1101/700757
Guo, D., Yuan, C., Luo, Y., Chen, Y., Lu, M., Chen, G., Ren, G., Cui, C., Zhang, J. and An, D. (2020). Biocontrol of tobacco black shank disease (Phytophthora nicotianae) by Bacillus velezensis Ba168. Pesticide Biochemistry and Physiology 165, 1-10. https://doi.ora/10.1016/i.pestbp.2020.01.004
Hardoim, P. R., Hardoim, C. C., van Overbeek, L. S. and van Elsas, J. D. (2012). Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS One 7, e30438. http://dx.doi.org/10.1371/journal.pone.0030438
Jozefkowicz, C., Brambilla, S., Frare, R., Stritzler, M., Puente, M., Piccinetti, C., Soto, G. and Ayub, N. (2017). Microevolution rather than large genome divergence determines the effectiveness of legume-rhizobia symbiotic interaction under field conditions. Journal of Molecular Evolutlon, 85, 79-83. https://doi.org/10.1007/s00239-017-9808-6
Li, H., Qiu, Y, Yao, T., Ma, Y., Zhang, H. and Yang, X. (2020). Effects of PGPR microbial inoculants on the growth and soil properties of Avena sativa, Medicago sativa and Cucumis sativus seedlings. Soil and Tillage Research, 199, 104577. https://doi.org/10.1016/j. still.2020.104577
López, J. L., Álvarez, F, Príncipe, A., Salas, M. E., Lozano, M. J., Draghi, W. O., Jofré, E. and Lagares, A. (2018). Isolation, taxonomic analysis and phenotypic characterization of bacterial endophytes present in alfalfa (Medicago sativa) seeds. Journal of Biotechnology, 267, 55-62. https://doi.org/10.1016/j. ¡biotec.2017.12.020
López-López, A., Rogel, M. A., Ormeno-Orrillo, E., Martínez-Romero, J., and Martínez-Romero, E. (2010). Phaseolus vulgaris seed-borne endophytic community with novel bacterial species such as Rhizobium endophyticum sp nov. Systematic and Applied Microbiology, 33(6), 322-327. https://doi. orq/10.1016/j.syapm.2010.07.005
Malfanova, N., Lugtenberg, B. J. J. and Berg, G. (2013). Bacterial endophytes: who and where, and what are they doing there? In: F J. DeBruijn (Ed.), Molecular Microbial Ecology of the Rhizosphere (pp. 391-403). Wiley-Blackwell Hoboken. https://doi. org/10.1002/9781118297674.ch36
Mukhopadhyay, K., Garrison, N. K., Hinton, D. M., Bacon, C. W., Khush, G. S., Peck, H. D. and Datta, N. (1996). Identification and characterization of bacterial endophytes of rice. Mycopathologia, 134, 151-159. https://doi.org/10.1007/BF00436723
Noori, F., Etesami, H., Zarini, H. N., Khoshkholgh-Sima, N. A., Salekdeh, G. H. and Alishahi, F (2018). Mining alfalfa (Medicago sativa L.) nodules for salinity tolerant non-rhizobial bacteria to improve growth of alfalfa under salinity stress. Ecotoxicology and Environmental Safety, 162, 129-138. https://doi.org/10.1016/¡.ecoenv. 2018.06.092
Odorizzi, A. S. (2015). Parámetros genéticos, rendimiento y calidad forrajera en alfalfas (Medicago sativa L.) extremadamente sin reposo con expresión variable del carácter multifoliolado obtenidas por selección fenotípica recurrente. Tesis de doctorado. Facultad de Ciencias Agropecuarias, Universidad Nacional de Córdoba. Córdoba, Argentina. http://hdl.handle. net/11086/1834
Pini, F., Frascella, A., Santopolo, L., Bazzicalupo, M., Biondi, E. G., Scotti, C. and Mengoni, A. (2012). Exploring the plant-associated bacterial communities in Medicago sativa L. BMC Microbiology, 12, 78. http:// dx.doi.org/10.1186/1471-2180-12-78
Qiao, J., Wu, H., Huo, R., Gao, X. and Borriss, R. (2014). Stimulation of plant growth and biocontrol by Bacillus amyloliquefaciens subsp. plantarum FZB42 engineered for improved action. Chemical and Biological Technologies in Agriculture, 1, 12. https:// doi.org/10.1186/s40538-014-0012-2
Rodríguez, J., Ríos, Y. and Baró, Y. (2016). Efectividad de cepas de Azotobacter sp. y Bacillus sp. para el control de especies fúngicas asociadas a hortalizas. Cultivos Tropicales, 37, 13-19. http://scielo.sld.cu/scielo.php?pid = S0258-59362016000500002&script=sciarttext&tlng=en
Rojas-Badía, M. M., Sánchez Castro, D., Rosales Perdomo, K. and Lugo Moya, D. (2017). Antagonismo de Bacillus frente a hongos fitopatógenos de cultivos hortícolas. Revista de Protección Vegetal, 32(2), 1-9. http://scielo.sld.cu/scielo.php?pid=S1010-27522017000200005&script=sci arttext&tlng = pt
Ruzzi, M. and Aroca, R. (2015). Plant growth-promoting rhizobacteria act as biostimulants in horticulture. Scientia Horticulturae, 196, 124-134. http://dx.doi. org/10.1016/j.scienta.2015.08.042
Sundaramoorthy, S. and Balabaskar, P. (2013). Evaluation of combined efficacy of Pseudomonas fluorescens and Bacillus subtilis in managing tomato wilt caused by Fusarium oxysporum f. sp. lycopersici (Fol). Plant Pathology Journal, 12(4), 154-161. https://doi. org/10.3923/ppj.2013.154.161
Xu, M., Sheng, J., Chen, L., Men, Y., Gan, L., Guo, S. and Shen, L. (2014). Bacterial community compositions of tomato (Lycopersicum esculentum Mill.) seeds and plant growth promoting activity of ACC deaminase producing Bacillus subtilis (HYT-12-1) on tomato seedlings. World Journal of Microbiology and Biotechnology, 30, 835-845. https://doi.org/10.1007/ s11274-013-1486-y
Yu, Y., Gui, Y., Li, Z., Jiang, C., Guo, J., and Niu, D. (2022). Induced Systemic Resistance for Improving Plant Immunity by Beneficial Microbes. Plants, 11, 386. https://doi.org/10.3390/plants11030386